Inheritance at the cellular level rests on the fact that one progenitor cell produces two identical daughter cells. The chromosomes are replicated and then, for each chromosome, the resulting sister chromatids move away from one another to opposite poles of the mitotic spindle apparatus (and, ultimately, into the two corresponding daughter cells). Chromosomes are relatively diffuse throughout most of the cell cycle, with no individualized units visible without specialized labeling. However, sister segregation requires that the two chromatids evolve into short, compact, individualized units which, ultimately, lie side-by-side.
Two recent papers from the Kleckner laboratory (Chu L, Liang Z, Mukhina M, Fisher J, Vincenten N, Zhang Z, Hutchinson J, Zickler D, Kleckner N. The 3D Topography of Mitotic Chromosomes. Mol Cell. 2020 Sep 17;79(6):902-916.e6. doi: 10.1016/j.molcel.2020.07.002. Epub 2020 Aug 7. PMID: 32768407; PMCID: PMC7502541; Chu L, Liang Z, Mukhina MV, Fisher JK, Hutchinson JW, Kleckner NE. One-dimensional spatial patterning along mitotic chromosomes: A mechanical basis for macroscopic morphogenesis. Proc Natl Acad Sci U S A. 2020 Oct 13:202013709. doi: 10.1073/pnas.2013709117. Epub ahead of print. PMID: 33051295) provide new insights which, together, reset the conversation regarding the fundamental nature of this individualization and compaction process. An important feature of these studies is that they rest on imaging of chromosomes in living cells and thus are not afflicted by the doubts and concerns of validity that have long plagued previous conclusions as drawn from analysis of fixed chromosomes.
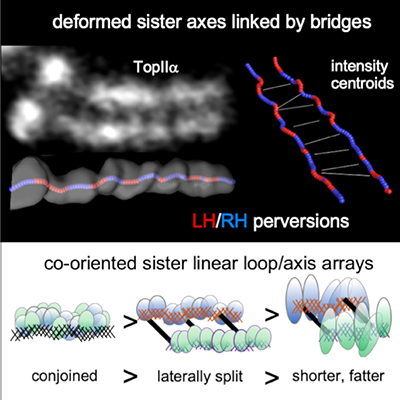
For 150 years, classical models have envisioned that chromosomes compact through a process that involves helical coiling. However, such models raise a conundrum: throughout the compaction process, sister chromatids lie side-by-side in parallel, with interactions at their shared interface. How could each individual chromatid undergo helical coiling while the sister pair maintains this parallel alignment? Chu et al. now provide the answer to this conundrum: in fact, contrary to dogma, mitotic chromosome compaction does not involve helical coiling but instead occurs by a topologically simple, linear process. In a previous study (Liang et al., 2015) the Kleckner lab showed that, just prior to onset of compaction, mitotic chromosomes comprise a morphologically single unit, with sister chromatids tightly linked in parallel as cooriented arrays of loops emanating from a conjoined structural axis. In their study in Molecular Cell, Chu et al. reveal that the parallel sister arrays first separate laterally and then progressively become shorter and fatter along their lengths. This compaction process likely involves restructuring of loop/axis arrays to give fewer, longer loops.
Chu et al.’s Molecular Cell study also revealed that sister chromatid axes are robustly linked by bridges, whose composition implies that they are “miniature axes”. Live cell imaging suggests that the fundamental role of these bridges is to keep sister chromatids in close parallel alignment despite disruptive forces due to the internal turbulence of compaction and the external turbulence that accompanies association of chromosomes with the developing mitotic spindle. In accord with this idea, bridges are quite evenly spaced, at a distance comparable to the perturbations to which they are subjected, thereby ensuring that they are effective in maintaining parallel alignment along the entire lengths of the chromosomes.
A third finding from this study is that chromosome axis paths, while linear, are not straight. Specifically, each axis comprises a sequential array of regular half-helical segments of alternating handedness. Such reversals of handedness are known as “perversions” and are common in both physical systems and in biological systems, most famously as first described by Darwin for vine tendrils. However, perversions have not previously been described for a sub-cellular object. Moreover, axis perversions have a complex structure which includes asymmetric bends and rotations at the positions of handedness changes. In retrospect, this unexpected axis path, having a tendency for helicity while being essentially linear, can explain why chromosome morphologies were mistakenly thought to be simple helices for so many years.
Together the above findings presented the authors with two new and interesting examples of spatial patterning in a biological system: evenly spaced bridges and regularly spaced deformations. Moreover, the periodicities of the two features are related (at ~400nm and ~200nm, respectively), pointing to a mechanistic relationship. Formation of any spatial pattern requires communication through the system. In a biological system, patterns may arise by a biochemical reaction-diffusion process, with communication provided by molecular diffusion. Alternatively, in physical systems and potentially also in biological systems, they may arise by a mechanical process, with communication provided by redistribution of mechanical stress.
A constellation of morphological and functional findings has led the Kleckner group, in collaboration with John Hutchinson of SEAS, to propose a mechanism of the latter type. By this model, mechanical stress accumulates within an axis, first leading to deformations and then driving emergence of bridges between axes. Bridges tend to appear at positions of handedness changes, which are the weakest points along the axis. The authors further propose that, after bridge emergence, continued presence of stress within axes creates a high energy state that enables the axis restructuring responsible for chromosome compaction.
Elucidation of the detailed basis for axis deformation and bridge emergence provides a unique and interesting new challenge which will require a combination of theoretical modeling and direct experimentation. The authors propose a model that invokes interplay between the bonded bilayer of axis and chromatin compartments in the context of classical theories of elastic rods from Kirchhoff and Love. Importantly, also, chromosome axes are not continuous proteinaceous filaments but rather are complex meshworks of proteins and catenated chromatin loops. The existence of a mechanical basis for the observed effects will provide a new perspective from which to understand the specific roles of involved molecules.
Finally, the two studies of Chu et al revealed direct, fundamental parallels between mitotic and meiotic chromosomes, as revealed by previous work from the Kleckner group. Together these findings offer promising insights into general chromosome dynamics and the evolutionary relation between the mitotic and meiotic programs.
by Nancy Kleckner






